We are a computational neuroscience group based at the University of Cambridge
The NeuroInformatics Research Group is a computational neuroscience group based in Department of Psychology at the University of Cambridge with a strong interest in studying lifespan brain health. A multitude of factors affect our brain health from as early as pre-conception until old age. These factors can pose great threats to the brain, leading to immense missed developmental potential, global disease burden and disability. We aim to tackle the diversity of challenges to studying brain health across the lifespan by using big data (both in terms of number of individuals, as well as in terms of numbers of features) and by developing novel approaches to integrate data across modalities and scales.
In order to understand atypical neurodevelopment or ageing, it is important to first understand typical development and ageing. To accomplish that our group develops life-spanning normative models for non-invasive brain imaging techniques such as MRI or CT akin to well know paediatric growth charts. We also use these normative reference models in a variety of applications. For example to study clinically relevant deviations, environmental factors influencing brain development and the genetic ontology of typical and atypical brain development.
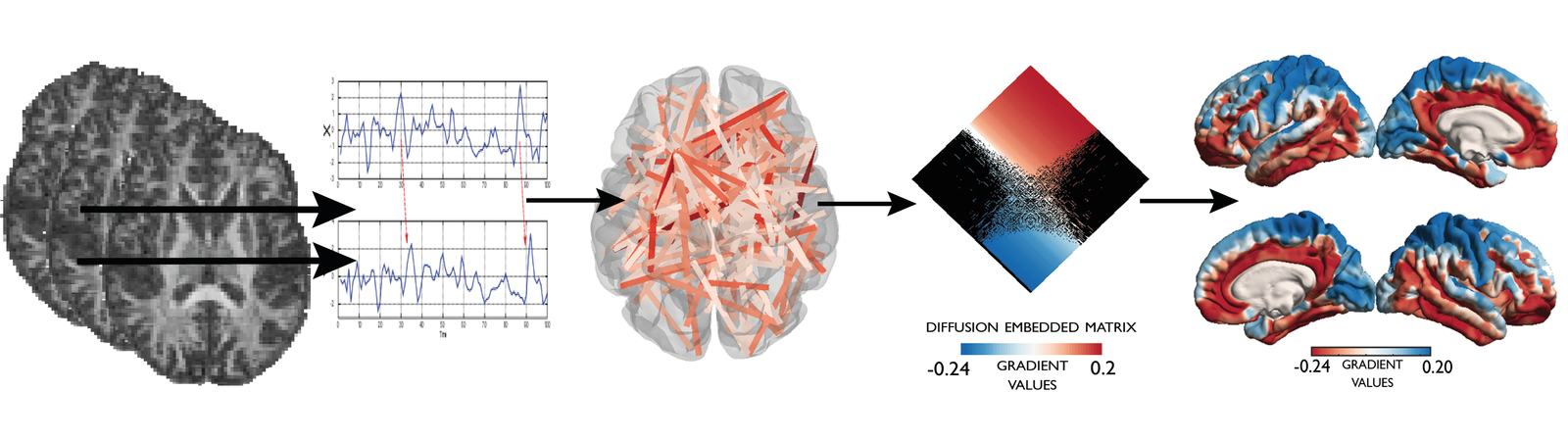
Big data does not always mean large samples but can also refer to large numbers of phenotypes. Especially when dealing with multi-modal or connectomic data the number of phenotypes can easily sky-rocket. Our group works on novel ways of integrating different levels and modalities of in-vivo imaging and in subsequently reducing their complexity through dimensionality reduction techniques such as graph theory and diffusion map embedding. We also explore the potential use of these lower dimensional representation for clinical stratification and subgrouping.
Emerging evidence from neuroimaging and genetics research has demonstrated that integrating genomic information from neuroimaging and neuropsychiatric phenotypes may aid in the identification of relevant genes and genetic pathways suitable for intervention. Consequently, there is a need to systematically identify shared genes and genetic processes underlying brain structure and function and neuropsychiatric conditions.
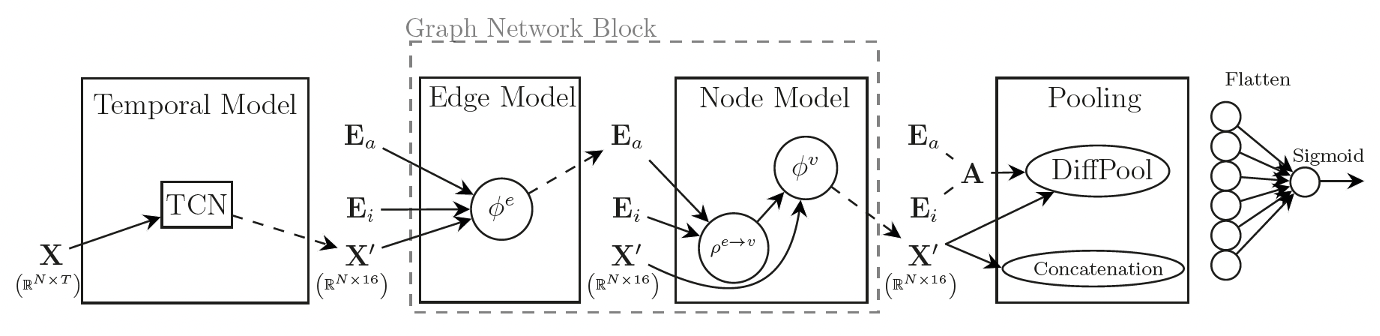
In an effort to take imaging and behavioural or phenotypic associations to the next level, our group, in close collaboration with Prof. Pietro Lio in the Computer Sciences department and Prof. Zoe Kourtzi in the Adaptive Brain Lab, also works on predictive modelling. That is, we try and build interpretable machine learning models that can be used for clinical stratification, prognostication and biomarker discovery.
Publication highlights
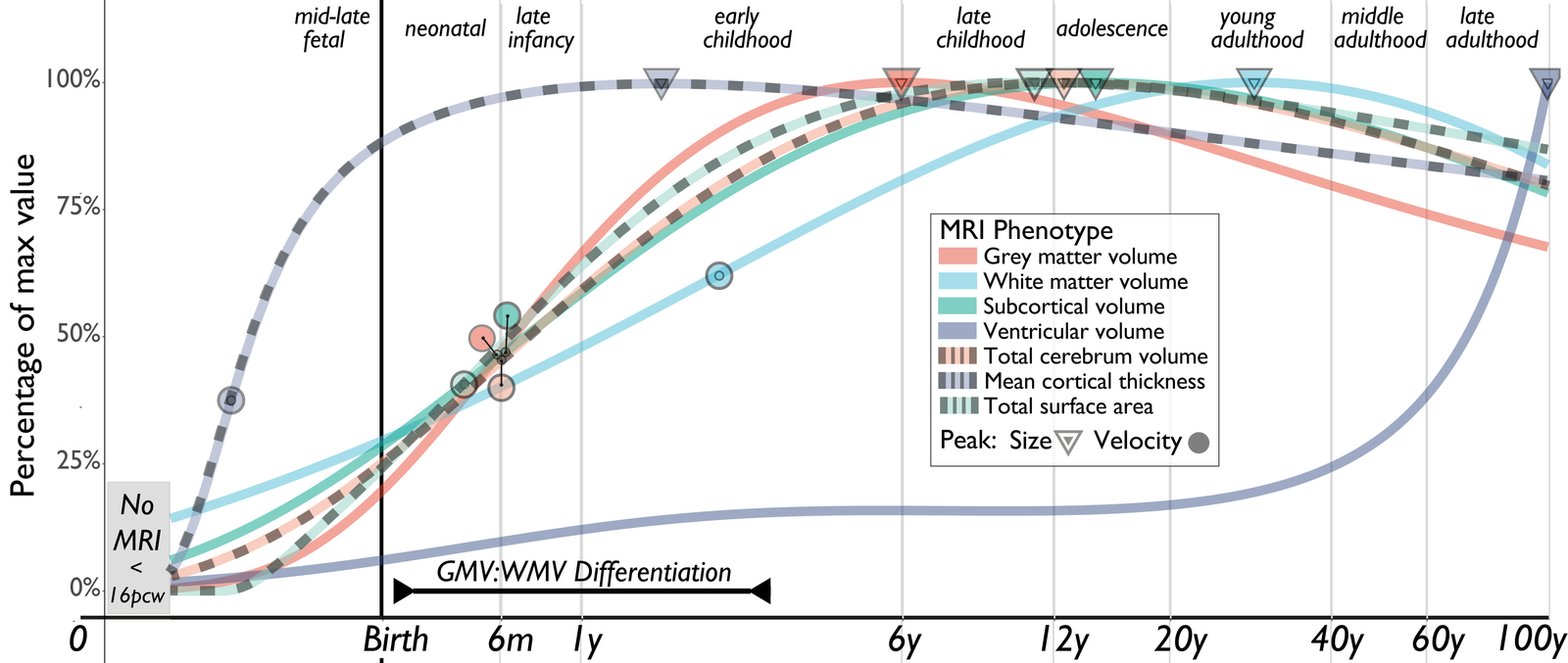
Neuroimaging is widely used in the study of the human brain, but no standards exist for measuring individual differences over time. To address this, we created an open resource, brainchart.io, to analyze MRI data. We used 123,984 MRI scans from 101,457 individuals ranging from 115 days post-conception to 100 years old to create reference charts. These charts quantify differences in MRI metrics using centile scores and highlight developmental milestones, stability of the brain across time, and robustness to differences in primary studies.
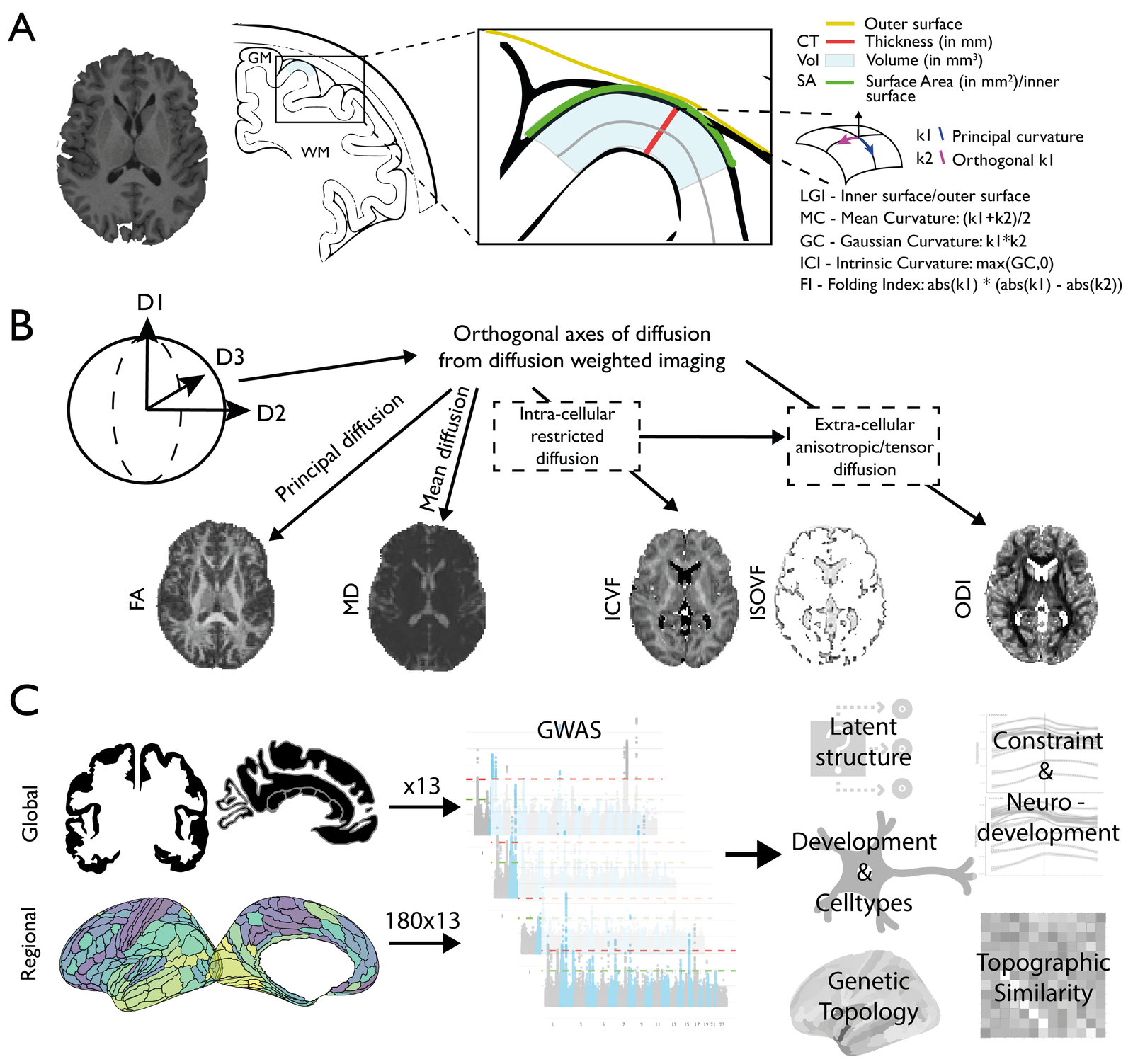
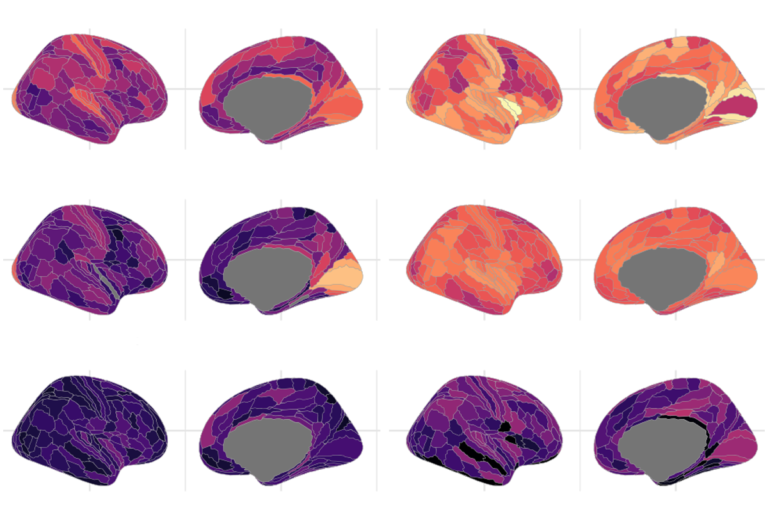
Our understanding of the genetics of the human cerebral cortex is limited both in terms of the diversity and the anatomical granularity of brain structural phenotypes. Here we conducted a genome-wide association meta-analysis of 13 structural and diffusion magnetic resonance imaging-derived cortical phenotypes, measured globally and at 180 bilaterally averaged regions in 36,663 individuals and identified 4,349 experiment-wide significant loci.
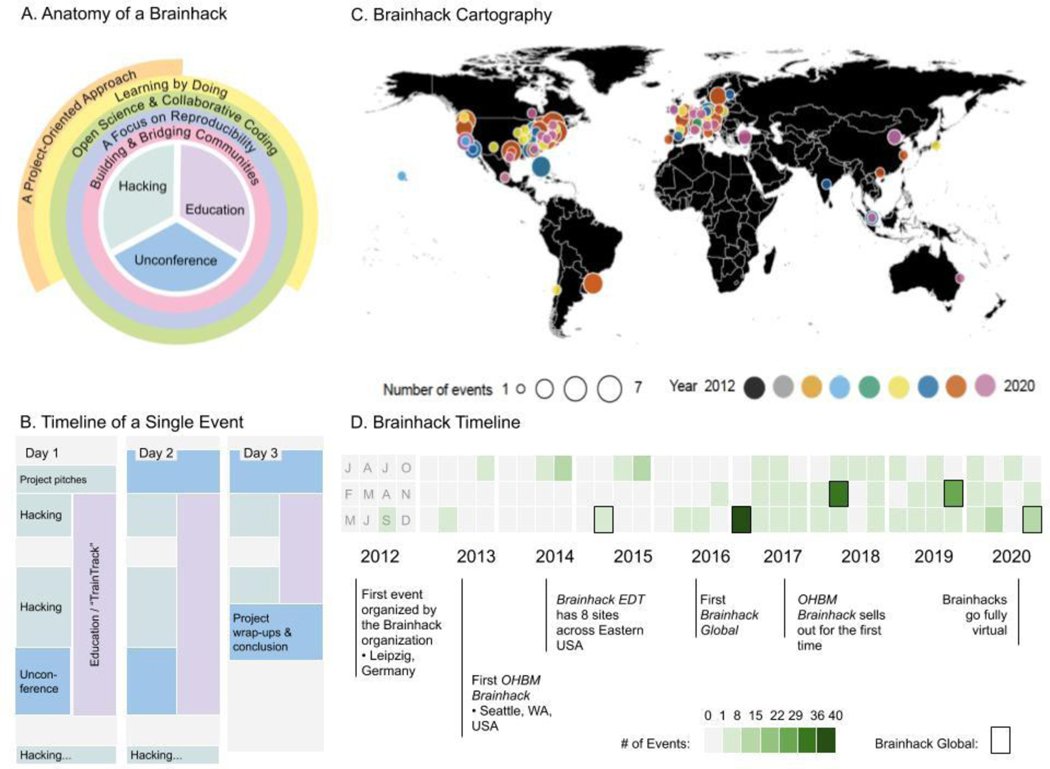
Brainhack is an innovative meeting format that promotes scientific collaboration and education in an open, inclusive environment. This NeuroView describes the myriad benefits for participants and the research community and how Brainhacks complement conventional formats to augment scientific progress.
Our research in the news
28 February 2023
11 March 2023
06 April 2022